神經生物實驗室
-

- 專長:Genetics, Neurobiology, Disease models
- 信箱:tzuyanglin@gate.sinica.edu.tw
- 電話:02-2787-1533
- 位置:R536/ICOB
Abstract
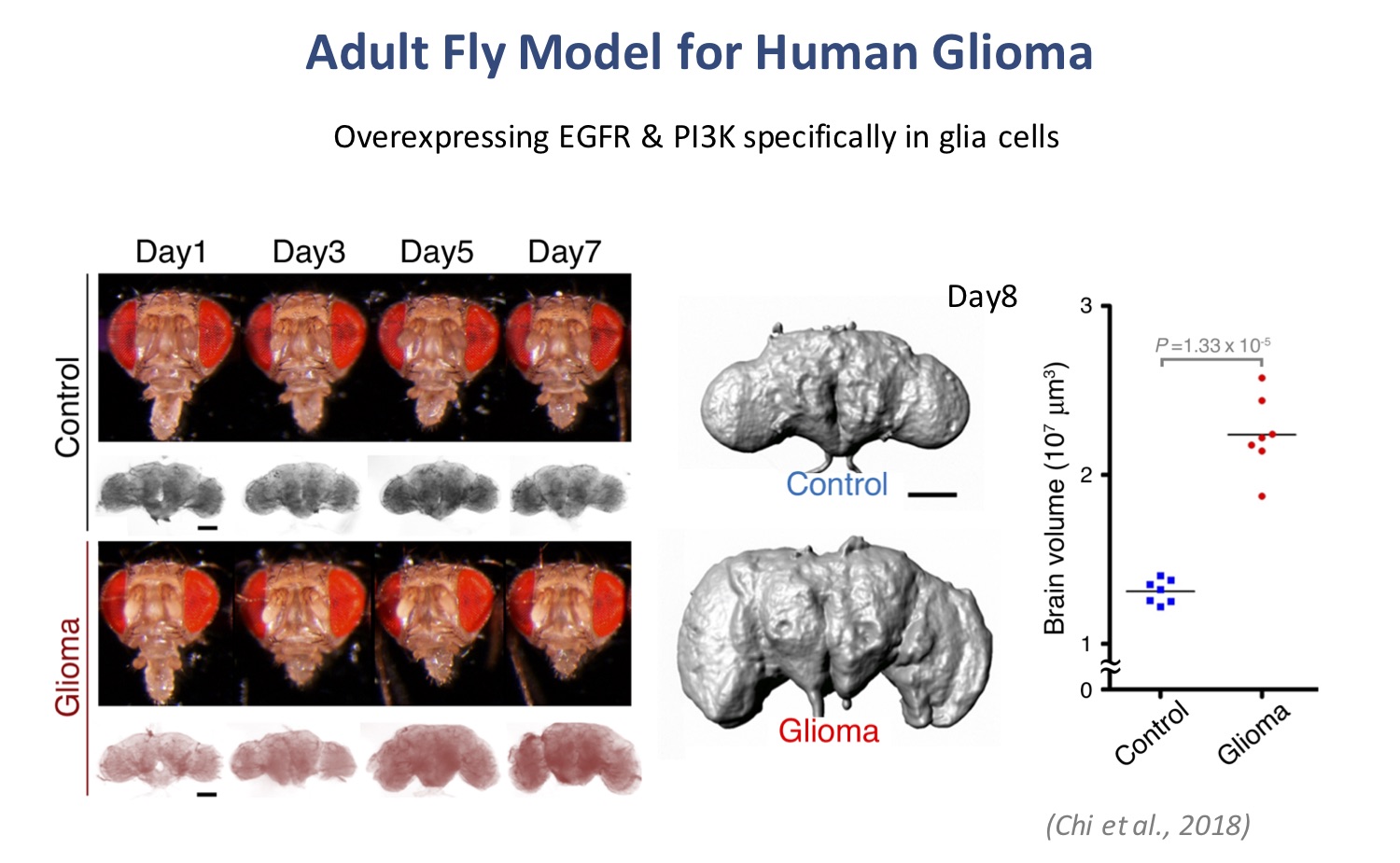
Glioblastoma multiforme (GBM), the most prevalent brain tumor in adults, has extremely poor prognosis. Frequent genetic alterations that activate epidermal growth factor receptor (EGFR) and phosphatidylinositol-3 kinase (P13K) signaling, as well as metabolic remodeling, have been associated with gliomagenesis. To establish a whole-animal approach that can be used to readily identify individual pathometabolic signaling factors, we induced glioma formation in the adult Drosophila brain by activating the EGFR-PI3K pathway. Glioma-induced animals showed significantly enlarged brain volume, early locomotor abnormalities, memory deficits, and a shorter lifespan. Combining bioinformatics analysis and glial-specific gene knockdown in the adult fly glioma model, we identified four evolutionarily conserved metabolic genes, including ALDOA, ACATI, ELOVL6, and LOX, that were involved in gliomagenesis. Silencing of ACAT1, which controls cholesterol homeostasis, reduced brain enlargement and increased the lifespan of the glioma-bearing flies. In GBM patients, ACAT1 is overexpressed and correlates with poor survival outcomes. Moreover, pharmacological inhibition of ACAT1 in human glioma cell lines revealed that it is essential for tumor proliferation. Collectively, these results imply that ACAT1 is a potential therapeutic target, and cholesterol homeostasis is strongly related to glioma formation. This in vivo model provides several rapid and robust phenotypic readouts, allowing determination of the pathometabolic pathways involved in gliomagenesis, as well as providing valuable information for novel therapeutic strategies.
2024 第21屆國家新創獎 (National Innovation Award)
| 姓名 | 職稱 | 電話 | 備註 | |
|---|---|---|---|---|
| 林子暘 | 研究副技師 | 2787-1533 | tzuyanglin@gate.sinica.edu.tw | |
| 鄭心瑜 | 研究助理 | 2787-1544 | shinyu1108@gate.sinica.edu.tw | |
| 陳姍渝 | 研究生(碩士) | 2787-1544 | chen0081@gate.sinica.edu.tw | 國防生化所 |
- Tian, X.J., Lin, T.Y., Lin, P.T., Tsai, M.J., Chen, W.J., Lee, C.M., Tu, C.H., Hsieh, T.H., Tung, Y.C., Wang, C.K., Lin, S., Chu, L.A., Tseng, F.G., Hsueh, Y.P., Lee, C.H., Chen, P., Chen, H., Hsu, J.C., and Chen, B.C. (2024) Rapid lightsheet fluorescence imaging of whole Drosophila brains at nanoscale resolution by potassium acrylate-based expansion microscopy. Nature Communications, 15, 10911 (2024). doi: 10.1038/s41467-024-55305-8
- Lin, C.H., Lin, T.Y., Hsu, S.C., and Hsu, H.C. (2022) Expansion Microscopy-based imaging for visualization of mitochondria in Drosophila ovarian germline stem cells. FEBS Open Bio 12 2102– 2110. doi: 10.1002/2211-5463.13506
- Li, Y., Chen, P.J., Lin, T.Y., Ting, C.Y., Muthuirulan, P., Pursley, R., Ilić, M., Pirih, P., Drews M.S., Menon, K.P., Zinn, K.G., Pohida, T., Borst, A., and Lee, C.H. (2021) Neural mechanism of spatio-chromatic opponency in the Drosophila amacrine neurons. Current Biology. 31 (14), 3040-3052, 2021-07. doi: 10.1016/j.cub.2021.04.068
- Lin, T. Y., Chen, P.J., Yu, H. H., Hsu, C. P., and Lee, C. H. (2021) Extrinsic Factors Regulating Dendritic Patterning. Front Cell Neurosci. Jan 13;14:622808. doi: 10.3389/fncel.2020.622808
-
Chen, P. L., Huang, K. T., Cheng, C. Y., Li, J. C., Chan, H. Y., Lin, T. Y., Su, M. P., Yang, W. Y., Chang, H. C., Wang, H. D., and Chen, C. H. (2020) Vesicular transport mediates the uptake of cytoplasmic proteins into mitochondria in Drosophila melanogaster. Nat Commun 11, 2592 doi: 10.1038/s41467-020-16335-0
-
Chakraborty, A, Lin, W. C., Lin, Y. T., Huang, K. J., Wang, P. Y., Chang, Y. F., Wang, H. L., Ma, K. T., Wang, C. Y., Huang, X. R., Lee, Y. H., Chen, B. C., Hsieh, Y. J., Chien, K. Y., Lin, T. Y., Liu, J. L., Sung, L. Y., Yu, J. S., Chang, Y. S., and Pai, L. M. (2020) SNAP29 Mediates the Assembly of Histidine-induced CTP Synthase Filaments in Proximity to the Cytokeratin Network. Journal of Cell Science doi: 10.1242/jcs.240200
-
Luo, J., Ting, C.Y., Li, Y., McQueen, P. G., Lin, T. Y., Hsu, C. P., and Lee, C. H. (2020) Antagonistic Regulation by Insulin-like Peptide and Activin Ensures the Elaboration of Appropriate Dendritic Field Sizes of Amacrine Neurons. eLife 2020:9:e50568.
-
Chi, K. C., Tsai, W. C., Wu, C. L., Lin, T. Y.*, and Hueng, D. Y.* (2019) An Adult Drosophila Glioma Model for Studying Pathometabolic Pathways of Gliomagenesis. Molecular Neurobiology, 56(6), 4589-4599. (* Corresponding Author)
-
Lin, W. C., Chakraborty, A, Huang, S. C., Wang, P. Y., Hsieh, Y. J., Chien, K. Y., Lee, Y. H., Chang, C. C., Tang, H. Y., Lin, Y. T., Tung, C. S., Luo, J. D., Chen, T. W., Lin, T. Y., Cheng, M. L., Chen, Y. T., Yeh, C. T., Liu, J. L., Sung, L. Y., Shiao, M. S., Yu, J. S., Chang, Y. S., and Pai, L. M. (2018) Histidine-Dependent Protein Methylation Is Required for Compartmentalization of CTP Synthase. Cell Reports, 24(10), 2733-2745.
-
Lin, T. Y., Luo, J., Shinomiya, K., Ting, C. Y., Lu, Z., Meinertzhagen, I.A., and Lee, C. H. (2016) Mapping Chromatic Pathways in the Drosophila Visual System. Journal of Comparative Neurology, 524(2), 213-227. (Featured cover image)
-
Macpherson, L.J., Zaharieva, E.E., Kearney, P.J., Alpert, M.H., Lin, T. Y., Turan, Z., Lee, C. H., and Gallio, M. (2015) Dynamic, Multi-color Labeling of Neural Connections by Trans-synaptic Fluorescence Complementation. Nature Communications, 6, doi:10.1038/ncomms10024
-
Miyazaki, T., Lin, T. Y., Ito, K., Lee, C. H., and Stopfer, M. (2015) A Gustatory Second-order Neuron that Connects Sucrose-sensitive Primary Neurons and a Distinct Region of the Gnathal Ganglion in the Drosophila Brain. Journal of Neurogenetics, 29(2-3), 144-155.
-
Karuppudurai, T.*, Lin, T. Y.*, Ting, C. Y., Pursley, R., Melnattur, K.V., Diao, F., White, B.H., Macpherson, L.J., Gallio, M., Pohida, T., and Lee, C. H. (2014) A Hard-wired Glutamatergic Circuit Pools and Relays UV Signals to Mediate Spectral Preference in Drosophila. Neuron, 81, 603-615. (*co-first author) (Recommended in F1000Prime)
-
Ting, C. Y., McQueen, P.G., Pandya, N., Lin, T. Y., Yang, M., Onteddu, V.R., O’Connor, M.B., McAuliffe, M., and Lee, C. H. (2014) Photoreceptor-derived dActivin Promotes Dendritic Termination and Restricts Receptive Fields of First-order Interneurons in Drosophila. Neuron, 81, 830-846
-
Melnattur, K.V., Pursley, R., Lin, T. Y., Ting, C. Y., Smith, P.D., Pohida, T., and Lee, C. H. (2014) Multiple Redundant Medulla Projection Neurons Mediate Color Vision in Drosophila. Journal of Neurogenetics, 28(3-4), 374-388
-
Shinomiya, K., Karuppudurai, T., Lin, T. Y., Lu, Z., Lee, C. H., and Meinertzhagen, I.A. (2014) Candidate Neural Substrates for Off-edge Motion Detection in Drosophila. Current Biology, 10, 1062-70.
-
Kundu, M., Kuzin, A., Lin, T. Y., Lee, C. H., Brody, T., and Odenwald, WF. (2013) cis-Regulatory Complexity within a Large Non-Coding Region in the Drosophila Genome. PLoS One, 8, e60137.
-
Brody, T., Yavatkar, A.S., Kuzin, A., Kundu, M., Tyson, L.J., Ross. J., Lin, T. Y., Lee, C. H., Awasaki, T., Lee, T., and Odenwald. W.F. (2012) Use of a Drosophila genome-wide conserved sequence database to identify functionally related cis-regulatory enhancers. Dev Dyn, 241, 169-189.
-
Ting, C. Y., Gu, S., Guttikonda, S., Lin, T. Y., White, B.H., and Lee, C. H. (2011) Focusing transgene expression in Drosophila by coupling Gal4 with a novel split-LexA expression system. Genetics, 188, 229-233.
-
Lin, T. Y.*, Huang, C. H.*, Kao, H. H., Liou, G. G., Yeh, S. R., Cheng, C. M., Chen, M. H., Pan, R. L., and Juang, J. L. (2009) Abi plays an opposing role to Abl in Drosophila axonogenesis and synaptogenesis. Development, 136 (18), 3099-3107. (*co-first author)
-
Huang, C. H., Lin, T. Y., Pan, R. L., and Juang, J. L. (2007). The involvement of Abl and PTP61F in the regulation of Abi protein localization and stability and lamella formation in Drosophila S2 cells. J Biol Chem 282, 32442-52.
-
Lin, H.*, Lin, T. Y.*, and Juang, J. L. (2007). Abl deregulates Cdk5 kinase activity and subcellular localization in Drosophila neurodegeneration. Cell Death & Differentiation 14 (3), 607-615. (*co-first author)
-
Chen, C. W., Lin, T. Y., Chen, C. T., and Juang, J. L. (2005). Distinct translation regulation by two alternative 5 ' UTRs of a stress-responsive protein - dPrx I. J Biomed Sci 12 (5), 729-739.
-
Lin, T. Y., Huang, C. H., Chou, W. G., and Juang, J. L. (2004). Abi enhances Abl-mediated CDC2 phosphorylation and inactivation. J Biomed Sci 11, 902-10.
-
Chiu, C. F., Lin, T. Y., and Chou, W. G. (2001). Direct transfer of Ku between DNA molecules with nonhomologous ends. Mutation Research/DNA Repair 486 (3), 185-194.
-
Roder, K.*, Hung, M. H.*, Lee, T. L.*, Lin, T. Y.*, Xiao, H., Isobe, K.I., Juang, J. L., and Shen, C. K. J. (2000). Transcriptional Repression by Drosophila Methyl-CpG-Binding Proteins. Molecular and cellular biology 20 (19), 7401-7409. (*co-first author)

