iTARGEX analysis of yeast deletome reveals novel regulators of transcriptional buffering in S phase and protein turnover
- Author:Dr. Jia-Hsin Huang, Ms.You-Rou Liao
- Journal: Nucleic Acid Research https://doi.org/10.1093/nar/gkab555
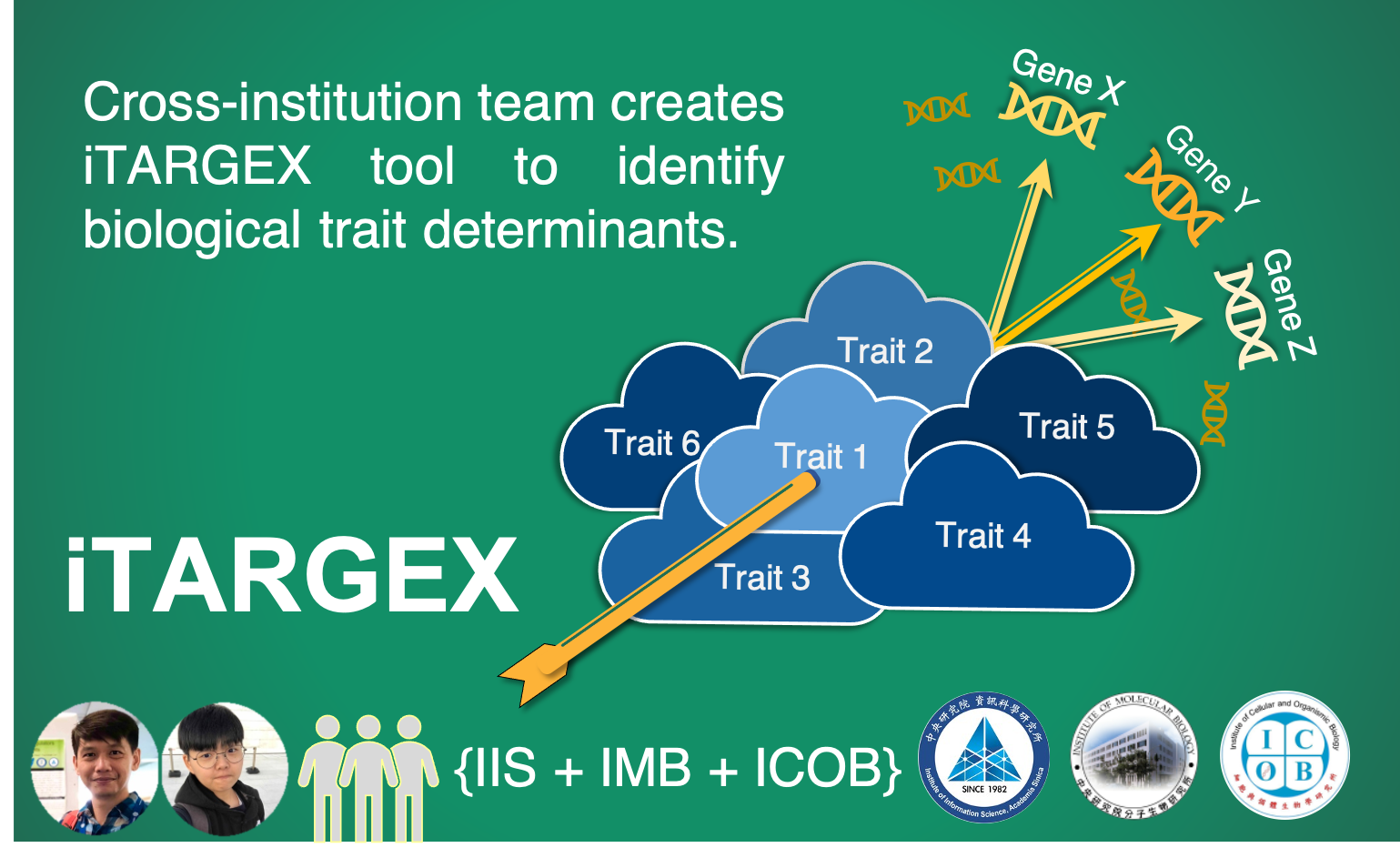
Cross-institution team creates “iTARGEX“ to identify biological trait determinants
Trait is a specific characteristic of an individual. Importantly, traits can be passed from one generation to another. For example, our hair color, blood type or height. Traits are determined by a gene or a group of genes, and they are determined by the interaction with the environment with genes. We often hear the word "phenotype", which is sometimes used interchangeably with the word “trait”, but "phenotype" may also define a whole compendium of traits together. There is consistently intense interest in the genetic factors contributing to many diseases, such as atherosclerosis, diabetes, and hypertension. These diseases are referred to as complex traits because multiple genes contribute to the phenotype either individually or through interactions with each other or the environment. However, it has been difficult in identifying potential determinants of phenotypic expression of the diseases, and this most often can be attributed to limitations in research tools. Integration of new efficient technologies for genotyping and public databases describing the fine structure of biological traits in the organisms should aid novel aspects of the gene discovery process.
To meet this need, our research team creates iTARGEX, a new bioinformatics tool, for in silico prediction of regulators linked to biological traits. Utilizing publicly available transcriptomic data from genetic perturbation experiments in the budding yeast, we successfully applied iTARGEX to identify novel regulators of gene expression homeostasis during S phase and global protein turnover. The functions of the novel regulators were validated experimentally. Notably, the novel regulators identified by iTARGEX have not been previously annotated with these biological functions. Therefore, our tool can facilitate the discovery of novel regulators by integration of omics and functional data. As a result, researchers can further generate specific novel hypotheses from the list of candidate genes that might not be easily connected through the conventional molecular biology approaches. The concept of iTARGEX provides an alternative approach to use of available biological data and lays a foundation for disease trait determinants.
-

The first author Dr. Jia-Hsin Huang -

The second author Ms.You-Rou Liao
The first author Dr. Jia-Hsin Huang, who was a postdoctoral fellow at Dr. Huai-Kuang Tsai’s laboratory in the Institute of Informatic Sciences, was responsible for the creation of iTARGEX. The second author Ms.You-Rou Liao, who was responsible for the experimental validation of the biological trait determinants predicted by iTARGEX, is a research assistant at Dr. Cheng-Fu Kao’s laboratory, Institute of Cellular and Organismic Biology. Research groups that collaborated on this project include those headed by Dr. Jun-Yi Leu at the Institute of Molecular Biology. This study was supported by grants from the Academia Sinica.
This study was published in Nucleic Acid Research, one of the best journals in BIOCHEMISTRY & MOLECULAR BIOLOGY (IF=16.971).
Article title: “iTARGEX analysis of yeast deletome reveals novel regulators of transcriptional buffering in S phase and protein turnover”
The full article is available at: https://doi.org/10.1093/nar/gkab555