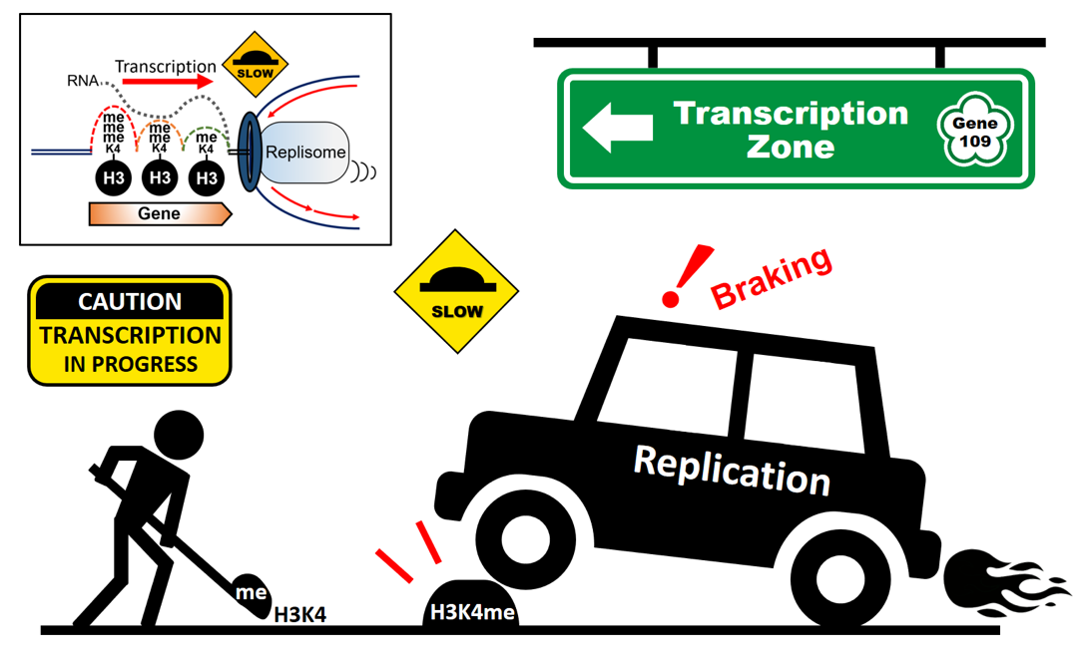
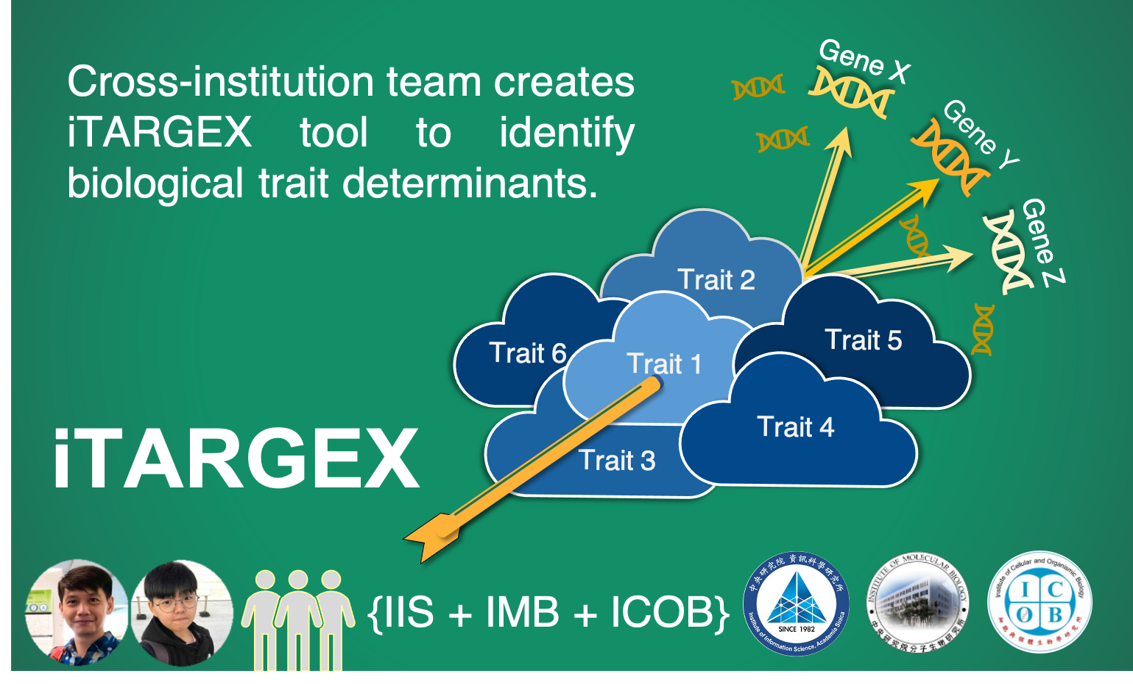
文章內容區
-

- 專長:Chromatin Structure & Dynamics, DNA Replication and Repair
- 信箱:ckao@gate.sinica.edu.tw
- 電話:02-2787-1515
- 網站: 高承福老師實驗室
- 位置:R243/ICOB