iTARGEX analysis of yeast deletome reveals novel regulators of transcriptional buffering in S phase and protein turnover
- 作者:Dr. Jia-Hsin Huang, Ms.You-Rou Liao
- 期刊: Nucleic Acid Research https://doi.org/10.1093/nar/gkab555
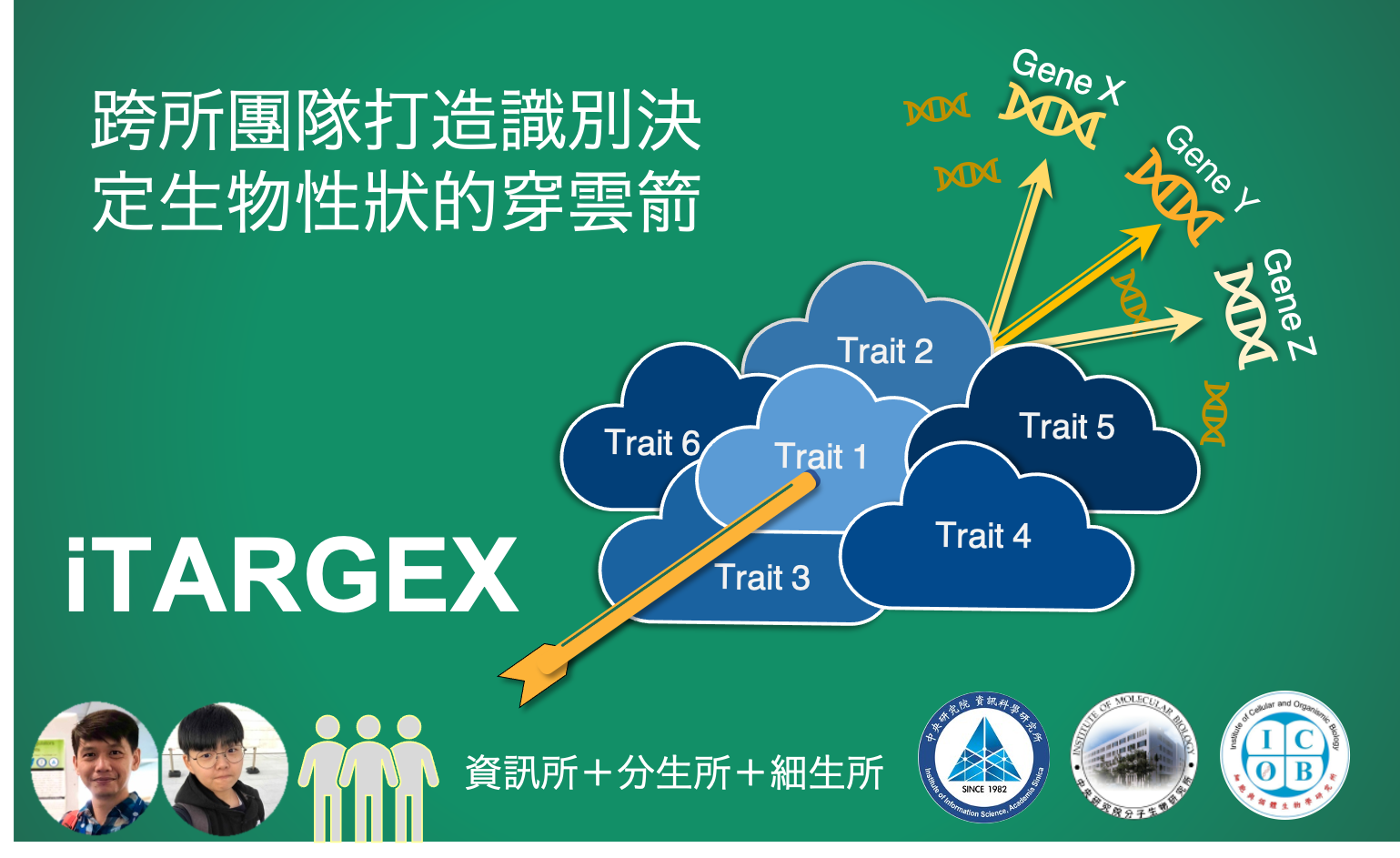
Cross-institution team creates “iTARGEX“ to identify biological trait determinants
Trait is a specific characteristic of an individual. Importantly, traits can be passed from one generation to another. For example, our hair color, blood type or height. Traits are determined by a gene or a group of genes, and they are determined by the interaction with the environment with genes. We often hear the word "phenotype", which is sometimes used interchangeably with the word “trait”, but "phenotype" may also define a whole compendium of traits together. There is consistently intense interest in the genetic factors contributing to many diseases, such as atherosclerosis, diabetes, and hypertension. These diseases are referred to as complex traits because multiple genes contribute to the phenotype either individually or through interactions with each other or the environment. However, it has been difficult in identifying potential determinants of phenotypic expression of the diseases, and this most often can be attributed to limitations in research tools. Integration of new efficient technologies for genotyping and public databases describing the fine structure of biological traits in the organisms should aid novel aspects of the gene discovery process.
To meet this need, our research team creates iTARGEX, a new bioinformatics tool, for in silico prediction of regulators linked to biological traits. Utilizing publicly available transcriptomic data from genetic perturbation experiments in the budding yeast, we successfully applied iTARGEX to identify novel regulators of gene expression homeostasis during S phase and global protein turnover. The functions of the novel regulators were validated experimentally. Notably, the novel regulators identified by iTARGEX have not been previously annotated with these biological functions. Therefore, our tool can facilitate the discovery of novel regulators by integration of omics and functional data. As a result, researchers can further generate specific novel hypotheses from the list of candidate genes that might not be easily connected through the conventional molecular biology approaches. The concept of iTARGEX provides an alternative approach to use of available biological data and lays a foundation for disease trait determinants.
-

The first author Dr. Jia-Hsin Huang -

The second author Ms.You-Rou Liao
The first author Dr. Jia-Hsin Huang, who was a postdoctoral fellow at Dr. Huai-Kuang Tsai’s laboratory in the Institute of Informatic Sciences, was responsible for the creation of iTARGEX. The second author Ms.You-Rou Liao, who was responsible for the experimental validation of the biological trait determinants predicted by iTARGEX, is a research assistant at Dr. Cheng-Fu Kao’s laboratory, Institute of Cellular and Organismic Biology. Research groups that collaborated on this project include those headed by Dr. Jun-Yi Leu at the Institute of Molecular Biology. This study was supported by grants from the Academia Sinica.
This study was published in Nucleic Acid Research, one of the best journals in BIOCHEMISTRY & MOLECULAR BIOLOGY (IF=16.971).
Article title: “iTARGEX analysis of yeast deletome reveals novel regulators of transcriptional buffering in S phase and protein turnover”
The full article is available at: https://doi.org/10.1093/nar/gkab555
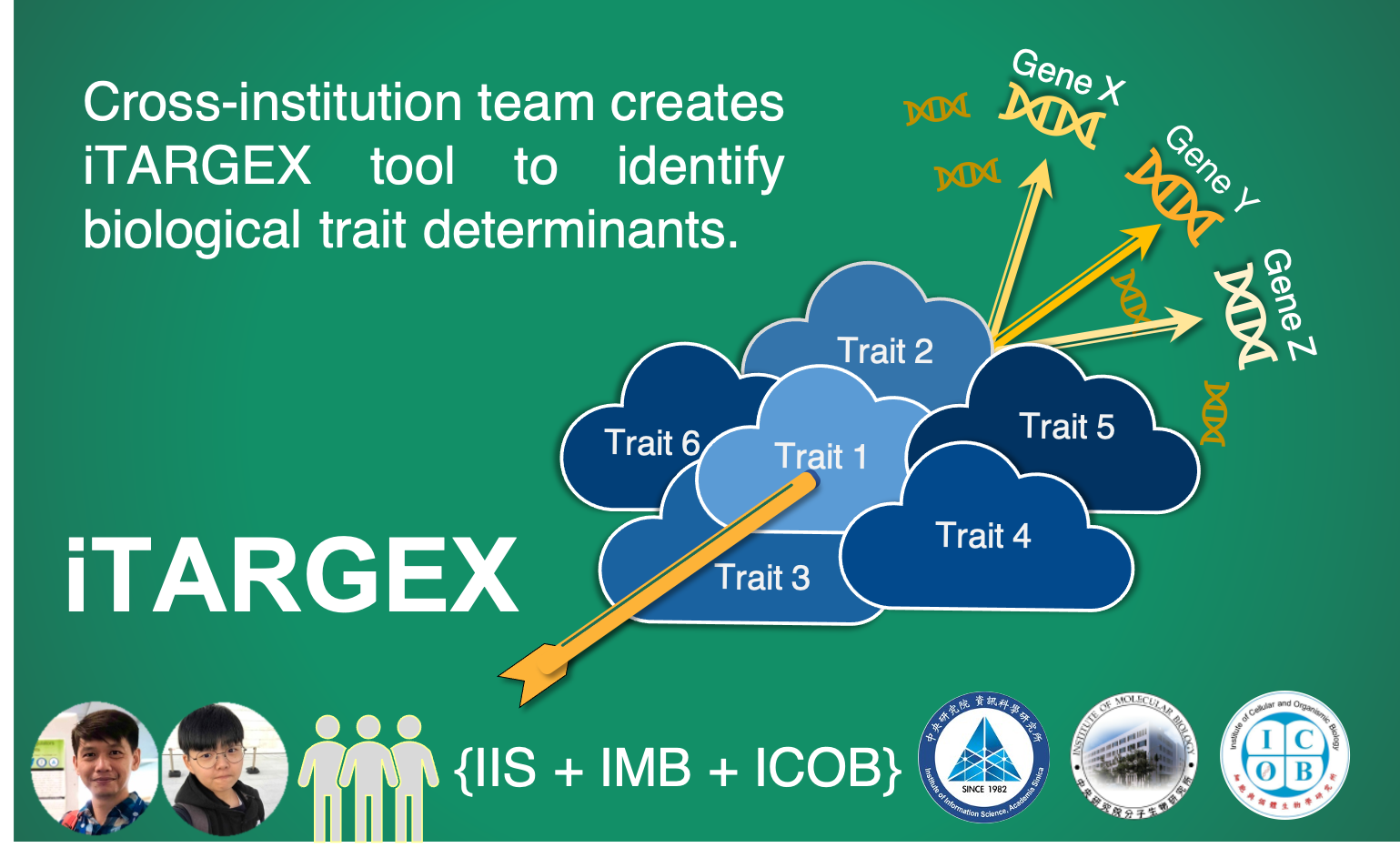
跨所團隊打造識別「生物性狀決定基因」的穿雲箭
性狀是生物體的的特定單一特徵。例如,我們的頭髮顏色、血型或身高。重要的是性狀可以代代相傳。性狀是由單一基因或是一組基因所決定的,也可由基因與環境的交互作用決定。 我們常聽到“表現型”這個詞與性狀通常是可以互換使用的,但“表現型”也可以用來定義一組完整的性狀。人們一直對導致許多疾病的遺傳基因有濃厚的興趣,例如動脈粥樣硬化、糖尿病和高血壓。這些疾病被稱為複雜性狀,因為多個基因單獨或通過彼此或環境的相互作用對表現型做出貢獻。找尋影響表現型的調控基因經常需要透過繁瑣的生物實驗與試誤的經驗累積。然而,隨著近年來生物技術應用的蓬勃發展,大規模的基因篩選與定序實驗以及生物性狀的描述研究已經累積充沛的資料,不過尚未有一套方法能夠整合不同生物性資料的資訊來找尋可能影響某個性狀的調控基因。
為了滿足這個需求,我們的研究團隊打造了 「iTARGEX」,一個全新的生物資訊工具,從剔除單一基因後的整體基因表現量變化與生物特徵進行一系列的相關性分析去預測參與調節特定性狀的基因。利用來自酵母菌基因遺傳實驗的公開轉錄組數據,我們成功地應用 iTARGEX 來預測控制「細胞週期」和調整「細胞內蛋白質周轉率」的調節因子。我們團隊並透過生物實驗進一步驗證了新找到的調節因子在細胞中所扮演的功能。值得注意的是,iTARGEX 鑑定的新調節因子對於該性狀的調控功能大都是尚未被過去的研究或文獻所提及,也很困難用既有的基因功能註釋去聯想而得到。因此,我們的生物資訊工具可以幫助研究人員對所找到的候選基因去生成全新的假說,突破舊有的知識框架,拓展對性狀調控機制的了解。此外,我們的工具通過整合多組學和基因功能數據來促進新調節因子的發現,亦可延伸應用於尋找不同疾病的特徵因子。
-

第一作者黃佳欣博士 -

第二作者廖宥媃小姐
負責創建iTARGEX的第一作者黃佳欣為資訊科學研究所蔡懷寬博士實驗室的博士後研究員。第二作者廖宥媃,負責實驗驗證iTARGEX所預測的生物性狀決定因子,是細胞與生物研究所高承福博士實驗室的研究助理。參與該項目的研究小組還包括由分子生物學研究所的呂俊毅博士領導的研究團隊。這項研究由中央研究院的主題計畫所資助。本研究及其結果發表在Nucleic Acid Research,是生化與分生領域最好的期刊之一 (IF=16.971)。
論文全文:“iTARGEX analysis of yeast deletome reveals novel regulators of transcriptional buffering in S phase and protein turnover” (https://doi.org/10.1093/nar/gkab555)