Molecular asymmetry in the cephalochordate embryo revealed by single-blastomere transcriptome profiling
- 作者:Che-Yi Lin, Mei-Yeh Jade Lu, Jia-Xing Yue, Kun-Lung Li, Yann Le Pétillon, Luok Wen Yong, Yi-Hua Chen
- 期刊: PLoS Genetics (2020) 16(12): e1009294. doi: 10.1371/journal.pgen.1009294. https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009294
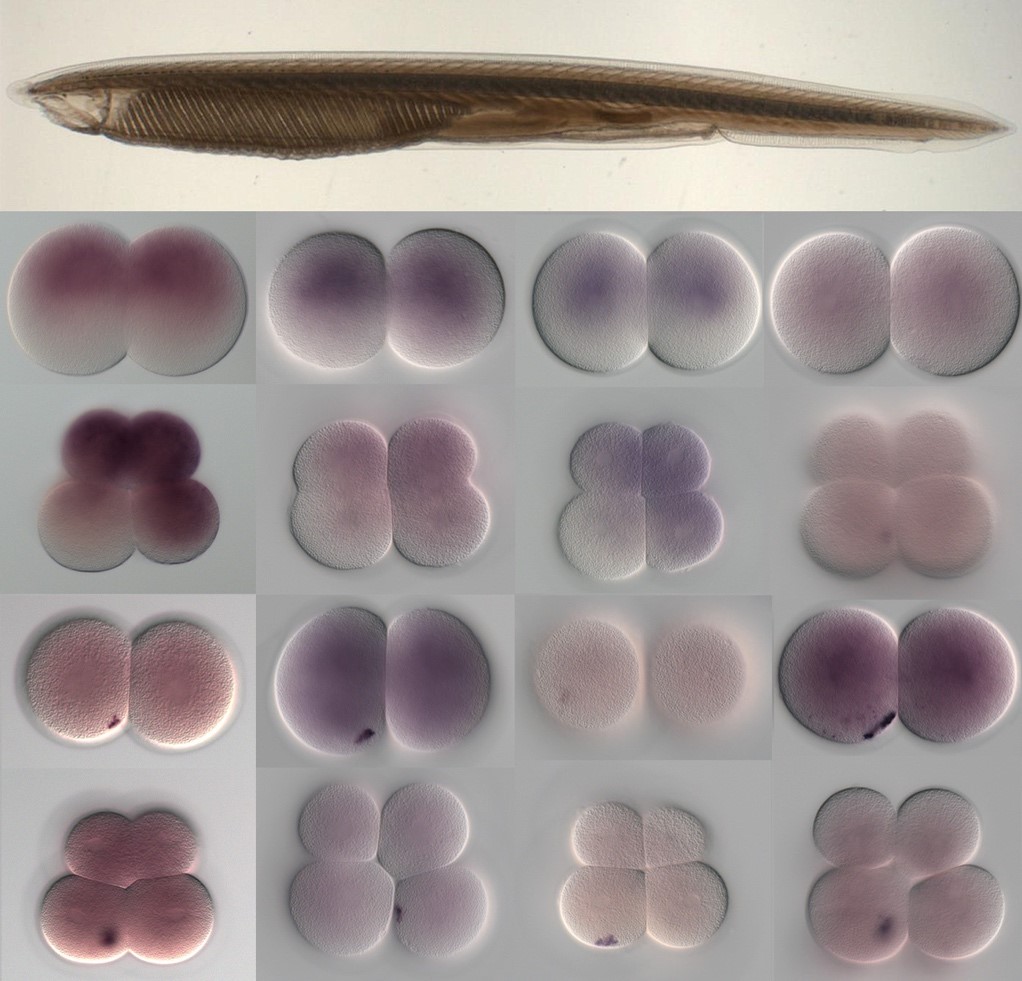
Studies in various animals have shown that asymmetrically localized maternal transcripts play important roles in axial patterning and cell fate specification in early embryos. However, comprehensive analyses of the maternal transcriptomes with spatial information are scarce and limited to a handful of model organisms. In cephalochordates (amphioxus), an early branching chordate group, maternal transcripts of germline determinants form a compact granule that is inherited by a single blastomere during cleavage stages. Further blastomere separation experiments suggest that other transcripts associated with the granule are likely responsible for organizing the posterior structure in amphioxus; however, the identities of these determinants remain unknown. In this study, we used high-throughput RNA sequencing of separated blastomeres to examine asymmetrically localized transcripts in two-cell and eight-cell stage embryos of the amphioxus Branchiostoma floridae. We identified 111 and 391 differentially enriched transcripts at the 2-cell stage and the 8-cell stage, respectively, and used in situ hybridization to validate the spatial distribution patterns for a subset of these transcripts. The identified transcripts could be categorized into two major groups: (1) vegetal tier/germ granule-enriched and (2) animal tier/anterior-enriched transcripts. Using zebrafish as a surrogate model system, we showed that overexpression of one animal tier/anterior-localized amphioxus transcript, zfp665, causes a dorsalization/anteriorization phenotype in zebrafish embryos by downregulating the expression of the ventral gene, eve1, suggesting a potential function of zfp665 in early axial patterning. Our results provide a global transcriptomic blueprint for early-stage amphioxus embryos. This dataset represents a rich platform to guide future characterization of molecular players in early amphioxus development and to elucidate conservation and divergence of developmental programs during chordate evolution.